概要
- 状態密度(Density Of States, DOS)を計算します
- 事前にSCF計算を行ってください
- TiCを例に説明します
手順
- DOSの計算には大量のk点数が必要です。k点数を増やします(最終的に得られる状態密度のグラフが収束するまで増やします)
$ x kgen NUMBER OF K-POINTS IN WHOLE CELL: (0 allows to specify 3 divisions of G) 10000 length of reciprocal lattice vectors: 1.342 1.342 1.342 21.544 21.544 21.544 286 k-points generated, ndiv= 21 21 21 KGEN ENDS 0.137u 0.004s 0:06.64 1.9% 0+0k 0+1376io 0pf+0w - x lapw1により電荷密度を計算します
$ x lapw1 -p
- スピン分極している場合は、アップスピンとダウンスピンの両方を計算する必要があります。以下、xによる計算が出てきた場合は同様です。出力ファイル名も最後にupあるいはdnが追加されます
$ x lapw1 -up -p $ x lapw1 -dn -p
- (必要ならば)case.inqを編集します。局所座標を回すことができます(気が向いたらその書き方も説明しようと思います)
- QTLというプログラムを使って軌道ごとの電荷密度を計算します
$ x qtl -p
- 以下のようにLAPW2でも軌道ごとの電荷密度が計算できますが、case.inqは読み込みません
$ x lapw2 -qtl -p
- case.qtl のヘッダーを見て、使用できる軌道を確認します。あるいは次のようにして確認します
$ grep "JATOM" TiC.qtl JATOM 1 MULT= 1 ISPLIT= 5 tot,s,p,d,d-eg,d-t2g, JATOM 2 MULT= 1 ISPLIT= 5 tot,s,p,
- DOSの計算の入力ファイル case.int を生成します。TiCの例です
$ configure_int_lapw -b total 1 tot,d,d-eg,d-t2g 2 tot,s,p end
- この場合は全DOS、1番目の原子 (Ti) のtot,d,d-eg,d-t2gの部分状態密度、2番目の原子 (C) のtot,s,pの部分状態密度を計算します。
- 最後の end を忘れがちなので気をつけてください。
- \$ configure_int_lapw だけで実行すれば対話形式で作成できます。
- 必要ならば case.int の2行目を編集します
- TiC.int
TiC #Title 0.25767 0.002 1.2492922854 0.003 #Emin, DE, Emax, Gauss-Broad 8 N 0.000 #Number of DOS-cases,G/L/B broadening (Ry) 0 1 total-DOS 1 1 tot-Ti 1 4 d-Ti 1 5 d-eg-Ti 1 6 d-t2g-Ti 2 1 tot-C 2 2 s-C 2 3 p-C SUM: 0 2 # NUMBER OF SUMMATIONS, max-nr-of summands 2 5 # this sums dos-cases 2+5 from the input above
- 2行目は状態密度の 下限、刻み幅、上限、ブロードニングの幅 を表します。単位はRyです。エネルギーは離散的に得られるので、式の上ではデルタ関数の和になってしまいますが、これをガウス関数で広がりをもったものにします
- case.intができたら、x tetraを実行します
$ x tetra -p
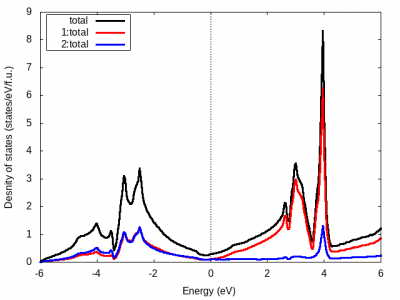
- 結果は case.dos1 と case.dos1ev に出力される(前者がエネルギー単位Ryで、後者がeV)ので、GNUPLOTなどでプロットします。
- 部分状態密度の軌道の種類が多いとcase.dos2, … に分割されます。